Recent Publications from the lab
-
Viral mimicry is a cellular state in which the reactivation of silenced transposable elements (TEs) leads to the accumulation of immunogenic nucleic acids, triggering innate immune pathways that resemble responses mounted against viral pathogens. Although they were first characterized in the context of epigenetic therapies, growing evidence indicates that other cancer treatment modalities – including radiotherapy, chemotherapies, and targeted therapies – can also induce TE reactivation and viral mimicry responses in cancer cells. This review synthesizes the current knowledge on treatment-induced TE-mediated immune responses in cancer, highlighting therapeutic strategies, shared and distinct molecular mechanisms, and their broader implications for tumor–immune interactions and treatment outcomes.
-
An emerging hallmark of many human diseases is transcription of typically silenced repetitive DNA containing pathogen-associated molecular patterns (PAMPs). These PAMPs engage the innate immune system via pattern recognition receptors (PRRs)—a phenomenon known as viral mimicry. We propose a statistical physics framework to quantify viral mimicry by measuring “selective forces” that enrich PAMPs compared to a genome-wide reference distribution. We validate our predictions by identifying repeats that bind different PRRs and show potential viral mimics in different repeat families across eukaryotic genomes, suggesting shared mechanisms drive emergence and retention. We propose two non-exclusive evolutionary hypotheses. The first “repeat-centric” hypothesis posits PAMPs are integral to the repeat life cycle and are therefore enriched as they mediate repeat expansion. The second “organism-centric” hypothesis proposes viral mimicry functions as a cell-intrinsic feedback mechanism for sensing and reacting to transcriptional dysregulation, which provides a selective pressure to maintain PAMPs in genomes.
-
mRNA vaccines have revolutionized vaccine development against infectious diseases, as demonstrated by the rapid creation and deployment of vaccines for SARS-CoV-2. In this chapter, we present a protocol for the generation of mRNA vaccine formulations utilizing lipid nanoparticle-based delivery systems. Additionally, we provide guidance for the evaluation of (i) mRNA formulation immunogenicity, (ii) mRNA formulation induced antigen cross-presentation, and (iii) antigen-specific CD8+ T-cell responses post in vivo vaccination.
-
Mismatch repair deficiency (MMRd), either due to inherited or somatic mutation, is prevalent in colorectal cancer (CRC) and other cancers. While anti-PD-1 therapy is utilized in both local and advanced disease, up to 50% of MMRd CRC fail to respond. Using animal and human models of MMRd, we determined that interactions between MHC+ C1Q+ CXCL9+ macrophages and TCF+ BHLHE40+ PRF1+ T cell subsets are associated with control of MMRd tumor growth, during anti-PD-1 treatment. In contrast, resistance is associated with upregulation of TIM3, LAG3, TIGIT, and PD-1 expression on T cells, and infiltration of the tumor with immunosuppressive TREM2+ macrophages and monocytes. By combining anti-PD-1 with anti-LAG3/CTLA4/TREM2, up to 100% tumor eradication was achieved in MMRd CRC and remarkably, in >70% in MMRp CRC. This study identifies key T cell and macrophage subsets mediating the efficacy of immunotherapy in overcoming immune escape in both MMRd and MMRp CRC settings.
-
Cell-intrinsic mechanisms of immunogenicity in ovarian cancer (OC) are not well understood. Damaging mutations in the SWI/SNF chromatin remodeling complex, such as SMARCA4 (BRG1), are associated with improved response to immune checkpoint blockade; however, the mechanism by which this occurs is unclear. We found that SMARCA4 loss in OC models resulted in increased cancer cell–intrinsic immunogenicity, characterized by up-regulation of long-terminal RNA repeats, increased expression of interferon-stimulated genes, and up-regulation of antigen presentation machinery. Notably, this response was dependent on STING, MAVS, and IRF3 signaling but was independent of the type I interferon receptor. Mouse ovarian and melanoma tumors with SMARCA4 loss demonstrated increased infiltration and activation of cytotoxic T cells, NK cells, and myeloid cells in the tumor microenvironment. These results were recapitulated in BRG1 inhibitor–treated SMARCA4-proficient tumor models, suggesting that modulation of chromatin remodeling through targeting SMARCA4 may serve as a strategy to overcome cancer immune evasion.
-
To thrive, cancer cells must navigate acute inflammatory signaling accompanying oncogenic transformation, such as via overexpression of repeat elements. We examined the relationship between immunostimulatory repeat expression, tumor evolution, and the tumor-immune microenvironment. Integration of multimodal data from a cohort of pancreatic ductal adenocarcinoma (PDAC) patients revealed expression of specific Alu repeats predicted to form double-stranded RNAs (dsRNAs) and trigger retinoic-acid-inducible gene I (RIG-I)-like-receptor (RLR)-associated type-I interferon (IFN) signaling. Such Alu-derived dsRNAs also anti-correlated with pro-tumorigenic macrophage infiltration in late stage tumors. We defined two complementary pathways whereby PDAC may adapt to such anti-tumorigenic signaling. In mutant TP53 tumors, ORF1p from long interspersed nuclear element (LINE)-1 preferentially binds Alus and decreases their expression, whereas adenosine deaminases acting on RNA 1 (ADAR1) editing primarily reduces dsRNA formation in wild-type TP53 tumors. Depletion of either LINE-1 ORF1p or ADAR1 reduced tumor growth in vitro. The fact that tumors utilize multiple pathways to mitigate immunostimulatory repeats implies the stress from their expression is a fundamental phenomenon to which PDAC, and likely other tumors, adapt.
-
As SARS-CoV-2 variants continue to emerge capable of evading neutralizing antibodies, it has become increasingly important to fully understand the breadth and functional profile of T cell responses to determine their impact on the immune surveillance of variant strains. Here, sampling healthy individuals, we profiled the kinetics and polyfunctionality of T cell immunity elicited by mRNA vaccination. Modeling of anti-spike T cell responses against ancestral and variant strains of SARS-CoV-2 suggested that epitope immunodominance and cross-reactivity are major predictive determinants of T cell immunity. To identify immunodominant epitopes across the viral proteome, we generated a comprehensive map of CD4+ and CD8+ T cell epitopes within non-spike proteins that induced polyfunctional T cell responses in convalescent patients. We found that immunodominant epitopes mainly resided within regions that were minimally disrupted by mutations in emerging variants. Conservation analysis across historical human coronaviruses combined with in silico alanine scanning mutagenesis of non-spike proteins underscored the functional importance of mutationally-constrained immunodominant regions. Collectively, these findings identify immunodominant T cell epitopes across the mutationally-constrained SARS-CoV-2 proteome, potentially providing immune surveillance against emerging variants, and inform the design of next-generation vaccines targeting antigens throughout SARS-CoV-2 proteome for broader and more durable protection.
Hematopoietic aging promotes cancer by fueling IL-1⍺–driven emergency myelopoiesis
Park MD, Le Berichel J, Hamon P, Wilk CM, Belabed M, Yatim N, Saffon A, Boumelha J, Falcomatà C, Tepper A, Hegde S, Mattiuz R, Soong BY, LaMarche NM, Rentzeperis F, Troncoso L, Halasz L, Hennequin C, Chin T, Chen EP, Reid AM, Su M, Reid Cahn A, Koekkoek LL, Venturini N, Wood-isenberg S, D’souza D, Chen R, Dawson T, Nie K, Chen Z, Kim-Schulze S, Casanova-Acebes M, Swirski FK, Downward J, Vabret N, Brown BD, Marron TU, Merad M. (2024) Science.
-
Age is a major risk factor for cancer, but how aging impacts tumor control remains unclear. Here, we establish that aging of the immune system, regardless of the age of the stroma and tumor, drives lung cancer progression. Hematopoietic aging enhances emergency myelopoiesis, resulting in the local accumulation of myeloid progenitor-like cells in lung tumors. These cells are a major source of IL-1⍺ that drives the enhanced myeloid response. The age-associated decline of DNMT3A enhances IL-1⍺ production, and disrupting IL-1R1 signaling early during tumor development normalized myelopoiesis and slowed the growth of lung, colonic, and pancreatic tumors. In human tumors, we identified an enrichment for IL-1⍺-expressing monocyte-derived macrophages linked to age, poorer survival, and recurrence, unraveling how aging promotes cancer and offering actionable therapeutic strategies.
Online immunology education for a global world
Vabret N, Mateus-Tique J , Salmon H, Martin JC, Mateo V, Serrano S, Lemoine F, Chammas R, Merad M. (2024) Nat Rev Immunol
Online technologies for immunology education can facilitate global outreach, incorporate local expertise, promote inclusivity and mitigate environmental costs of international travel. The experience gained from ten years of Immunoschool demonstrates how an engaging format with active participation is crucial to achieving these goals.
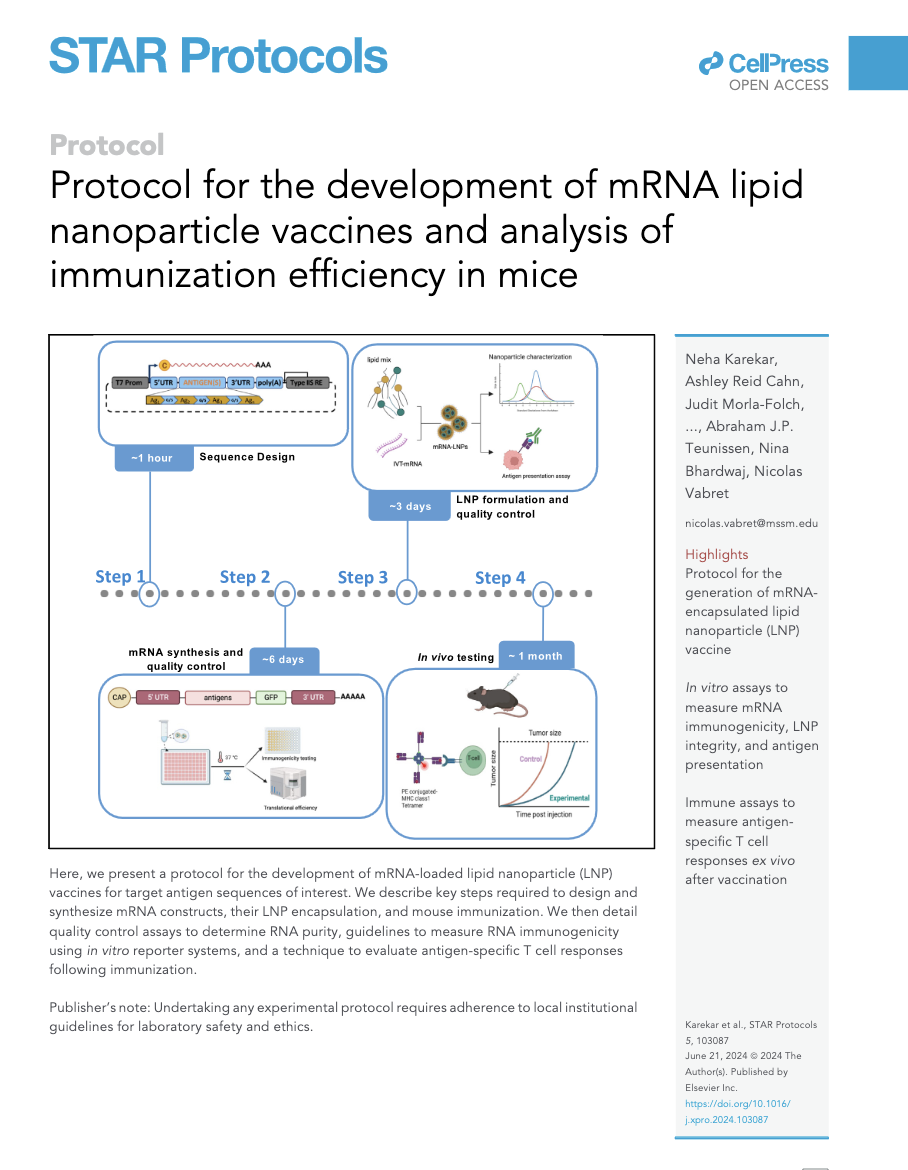
Protocol for the development of mRNA lipid nanoparticle vaccines and analysis of immunization efficiency in mice
Karekar N, Reid Cahn A , Morla-Folch J, , Saffon A , Ward RW, Ananthanarayanan A, Teunissen A, Bhardwaj N, Vabret N. (2024) STAR Protoc.
Here, we present a protocol for the development of mRNA-loaded lipid nanoparticle (LNP) vaccines for target antigen sequences of interest. We describe key steps required to design and synthesize mRNA constructs, their LNP encapsulation, and mouse immunization. We then detail quality control assays to determine RNA purity, guidelines to measure RNA immunogenicity using in vitro reporter systems, and a technique to evaluate antigen-specific T cell responses following immunization.
Featured in 2024 STAR Protocols Editors Collection: an editors' pick of the most notable papers (Top 5%) published in 2024.
Alternative RNA splicing generates shared clonal neoantigens across different types of cancer
Brown M, Vabret N. (2024) Nat Rev Immunol
Preprint clubs: why it takes a village to do peer review
Richter FC, Gea-Mallorquí E, Mortha A, Ruffin N, Vabret N. (2023) Nature Index
A group of early-career researchers have harnessed cross-institutional journal clubs to assess and review immunology preprints.
The Preprint Club - A blueprint for community-based peer review
Richter FC, Gea-Mallorquí E, Mortha A, Ruffin N, Vabret N. (2023) EMBO reports
Cross-institutional journal clubs focused on preprints are a new approach to community-based peer review and allow ERCs to gain experience.
The Preprint Club - A cross-institutional, community-based approach to peer reviewing
Richter FC, Gea-Mallorquí E, Ruffin N, Vabret N. (2023) Preprint at bioRxiv
-
The academic community has been increasingly using preprints to disseminate their latest research findings quickly and openly. This early and open access of non-peer reviewed research warrants new means from the scientific community to efficiently assess and provide feedback to preprints. Yet, most peer review of scientific studies performed today are still managed by journals, each having their own peer review policy and transparency. However, approaches to uncouple the peer review process from journal publication are emerging. Additionally, formal education of early career researchers (ECRs) in peer reviewing is rarely available, hampering the quality of peer review feedback. Here, we introduce the Preprint Club, a cross-institutional, community-based approach to peer reviewing, founded by ECRs from the University of Oxford, Karolinska Institutet and Icahn School of Medicine at Mount Sinai. Over the past two years and using the collaborative setting of the Preprint Club, we have been discussing, assessing, and providing feedback on recent preprints in the field of immunology. In this article, we provide a blueprint of the Preprint Club basic structure, demonstrate its effectiveness, and detail the lessons we learned on its impact on peer review training and preprint author's perception.
Dark genome, bright ideas: Recent approaches to harness transposable elements in immunotherapies
Reid Cahn A, Bhardwaj N, Vabret N. (2022) Cancer Cell
Transposable elements (TEs), which make up almost half of the human genome, often display altered expression in cancers. Here, we review recent progress in elucidating the role of TEs as mediators of immune responses in cancer and discuss how novel therapeutic strategies can harness TE immunogenicity for cancer immunotherapy.
Y RNAs are conserved endogenous RIG-I ligands across RNA virus infection and are targeted by HIV-1
Vabret N, Najburg V, Solovyov A, et al. (2022) iScience
-
Pattern recognition receptors (PRRs) protect against microbial invasion by detecting specific molecular patterns found in pathogens and initiating an immune response. Although microbial-derived PRR ligands have been extensively characterized, the contribution and relevance of endogenous ligands to PRR activation remains overlooked. Here, we characterize the landscape of endogenous ligands that engage RIG-I-like receptors (RLRs) upon infection by different RNA viruses. In each infection, several RNAs transcribed by RNA polymerase III (Pol3) specifically engaged RLRs, particularly the family of Y RNAs. Sensing of Y RNAs was dependent on their mimicking of viral secondary structure and their 5'-triphosphate extremity. Further, we found that HIV-1 triggered a VPR-dependent downregulation of RNA triphosphatase DUSP11 in vitro and in vivo, inducing a transcriptome-wide change of cellular RNA 5'-triphosphorylation that licenses Y RNA immunogenicity. Overall, our work uncovers the contribution of endogenous RNAs to antiviral immunity and demonstrates the importance of this pathway in HIV-1 infection.
Repeat elements amplify TLR signaling
O’Donnell T, Vabret N. (2022) Nat Rev Immunol
Highlight of Rookhuizen, D. C. et al. Induction of transposable element expression is central to innate sensing. Preprint at bioRxiv https://doi.org/10.1101/2021.09.10.457789 (2021)
Repeats Mimic Immunostimulatory Viral Features Across a Vast Evolutionary Landscape
Šulc P, Solovyov A, Marhon SA, Sun S, LaCava J, Abdel-Wahab O, Vabret N, De Carvalho DD, Monasson R, Cocco S, Greenbaum BD (2021) Preprint at bioRxiv
-
An emerging hallmark across many human diseases - such as cancer, autoimmune and neurodegenerative disorders – is the aberrant transcription of typically silenced repetitive elements. Once transcribed they can mimic pathogen-associated molecular patterns and bind pattern recognition receptors, thereby engaging the innate immune system and triggering inflammation in a process known as “viral mimicry”. Yet how to quantify pathogen mimicry, and the degree to which it is shaped by natural selection, remains a gap in our understanding of both genome evolution and the immunological basis of disease. Here we propose a theoretical framework that combines recent biological observations with statistical physics and population genetics to quantify the selective forces on virus-like features generated by repeats and integrate these forces into predictive evolutionary models. We establish that many repeat families have evolutionarily maintained specific classes of viral mimicry. We show that for HSATII and intact LINE-1 selective forces maintain CpG motifs, while for a set of SINE and LINE elements the formation of long double-stranded RNA is more prevalent than expected from a neutral evolutionary model. We validate our models by showing predicted immunostimulatory inverted SINE elements bind the MDA5 receptor under conditions of epigenetic dysregulation and that they are disproportionately present during intron retention when RNA splicing is pharmacologically inhibited. We conclude viral mimicry is a general evolutionary mechanism whereby genomes co-opt features generated by repetitive sequences to trigger the immune system, acting as a quality control system to flag genome dysregulation. We demonstrate these evolutionary principles can be learned and applied to predictive models. Our work therefore serves as a resource to identify repeats with candidate immunostimulatory features and leverage them therapeutically.
Highlights from a year in a pandemic
Merad M, Vabret N. (2021) J Exp Med
COVID-19 has emerged as one of the worst pandemics in recent history and has exposed the weaknesses of healthcare systems worldwide. Here, we reflect on the lessons learned from a year in a pandemic. We discuss the extraordinary scientific advances made in our understanding of a new disease, the failed and successful attempts to halt its progression, and the impact of the pandemic on the scientific discourse within the global community.
SARS-CoV-2: the many pros of targeting PLpro
McClain CB, Vabret N. (2020) Signal Transduct Target Ther.
Antibody responses to SARS-CoV-2 short-lived
Vabret N. (2020) Nat Rev Immunology
Preclinical data from SARS-CoV-2 mRNA vaccine
Vabret N. (2020) Nat Rev Immunology
Immunology of COVID-19: Current State of the Science
Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM; Sinai Immunology Review Project. (2020) Immunity
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected millions of people worldwide, igniting an unprecedented effort from the scientific community to understand the biological underpinning of COVID19 pathophysiology. In this Review, we summarize the current state of knowledge of innate and adaptive immune responses elicited by SARS-CoV-2 infection and the immunological pathways that likely contribute to disease severity and death. We also discuss the rationale and clinical outcome of current therapeutic strategies as well as prospective clinical trials to prevent or treat SARS-CoV-2 infection.
Hydroxychloroquine: small effects in mild disease
Levantovsky R, Vabret N. (2020) Nat Rev Immunology
Global Cancer Transcriptome Quantifies Repeat Element Polarization between Immunotherapy Responsive and T Cell Suppressive Classes
Solovyov A, Vabret N, Arora KS, Snyder A, Funt SA, Bajorin DF, Rosenberg JE, Bhardwaj N, Ting DT, Greenbaum BD. (2018) Cell Report
It has been posited that anti-tumoral innate activation is driven by derepression of endogenous repeats. We compared RNA sequencing protocols to assess repeat transcriptomes in The Cancer Genome Atlas (TCGA). Although poly(A) selection efficiently detects coding genes, most non-coding genes, and limited subsets of repeats, it fails to capture overall repeat expression and co-expression. Alternatively, total RNA expression reveals distinct repeat co-expression subgroups and delivers greater dynamic changes, implying they may serve as better biomarkers of clinical outcomes. We show that endogenous retrovirus expression predicts immunotherapy response better than conventional immune signatures in one cohort yet is not predictive in another. Moreover, we find that global repeat derepression, including the HSATII satellite repeat, correlates with an immunosuppressive phenotype in colorectal and pancreatic tumors and validate in situ. In conclusion, we stress the importance of analyzing the full spectrum of repeat transcription to decode their role in tumor immunity.
Sequence-Specific Sensing of Nucleic Acids
Vabret N, Bhardwaj N, Greenbaum BD. (2017) Trends Immunol
Innate immune cells are endowed with many nucleic acid receptors, but the role of sequence in the detection of foreign organisms remains unclear. Can sequence patterns influence recognition? In addition, how can we infer those patterns from sequence data? Here, we detail recent computational and experimental evidence associated with sequence-specific sensing. We review the mechanisms underlying the detection and discrimination of foreign sequences from self. We also describe quantitative approaches used to infer the stimulatory capacity of a given pathogen nucleic acid species, and the influence of sequence-specific sensing on host-pathogen coevolution, including endogenous sequences of foreign origin. Finally, we speculate how further studies of sequence-specific sensing will be useful to improve vaccine design, gene therapy and cancer treatment.